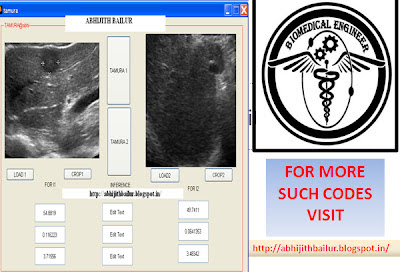
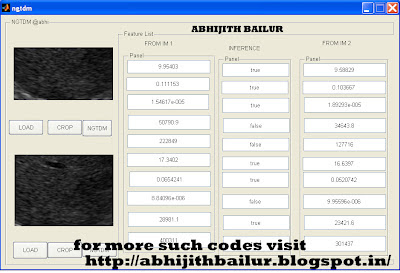
function varargout = tamura(varargin)
% ABHi TAMURA M-file for tamura.fig
% TAMURA, by itself, creates a new TAMURA or raises the existing
% singleton*.
%
% H = TAMURA returns the handle to a new TAMURA or the handle to
% the existing singleton*.
%
% TAMURA('CALLBACK',hObject,eventData,handles,...) calls the local
% function named CALLBACK in TAMURA.M with the given input arguments.
%
% TAMURA('Property','Value',...) creates a new TAMURA or raises the
% existing singleton*. Starting from the left, property value pairs are
% applied to the GUI before tamura_OpeningFcn gets called. An
% unrecognized property name or invalid value makes property application
% stop. All inputs are passed to tamura_OpeningFcn via varargin.
%
% *See GUI Options on GUIDE's Tools menu. Choose "GUI allows only one
% instance to run (singleton)".
%
% See also: GUIDE, GUIDATA, GUIHANDLES
% Edit the above text to modify the response to help tamura
% Last Modified by GUIDE v2.5 19-Jun-2012 00:35:01
% Begin initialization code - DO NOT EDIT
gui_Singleton = 1;
gui_State = struct('gui_Name', mfilename, ...
'gui_Singleton', gui_Singleton, ...
'gui_OpeningFcn', @tamura_OpeningFcn, ...
'gui_OutputFcn', @tamura_OutputFcn, ...
'gui_LayoutFcn', [] , ...
'gui_Callback', []);
if nargin && ischar(varargin{1})
gui_State.gui_Callback = str2func(varargin{1});
end
if nargout
[varargout{1:nargout}] = gui_mainfcn(gui_State, varargin{:});
else
gui_mainfcn(gui_State, varargin{:});
end
% End initialization code - DO NOT EDIT
% --- Executes just before tamura is made visible.
function tamura_OpeningFcn(hObject, eventdata, handles, varargin)
% This function has no output args, see OutputFcn.
% hObject handle to figure
% eventdata reserved - to be defined in a future version of MATLAB
% handles structure with handles and user data (see GUIDATA)
% varargin command line arguments to tamura (see VARARGIN)
handles.fileLoaded = 0;
handles.fileLoaded2=0;
set(handles.axes1,'Visible','off');
set(handles.axes2,'Visible','off');
% Choose default command line output for tamura
handles.output = hObject;
% Update handles structure
guidata(hObject, handles);
% UIWAIT makes tamura wait for user response (see UIRESUME)
% uiwait(handles.figure1);
% --- Outputs from this function are returned to the command line.
function varargout = tamura_OutputFcn(hObject, eventdata, handles)
% varargout cell array for returning output args (see VARARGOUT);
% hObject handle to figure
% eventdata reserved - to be defined in a future version of MATLAB
% handles structure with handles and user data (see GUIDATA)
% Get default command line output from handles structure
varargout{1} = handles.output;
% --- Executes on button press in LOAD1.
function LOAD1_Callback(hObject, eventdata, handles)
% hObject handle to LOAD1 (see GCBO)
% eventdata reserved - to be defined in a future version of MATLAB
% handles structure with handles and user data (see GUIDATA)
[FileName,PathName] = uigetfile({'*.*'},'Load Image File');
if (FileName==0) % cancel pressed
return;
end
handles.fullPath = [PathName FileName];
[a, b, Ext] = fileparts(FileName);
availableExt = {'.bmp','.jpg','.jpeg','.tiff','.png','.gif'};
FOUND = 0;
for (i=1:length(availableExt))
if (strcmpi(Ext, availableExt{i}))
FOUND=1;
break;
end
end
if (FOUND==0)
msgbox('File type not supported Load file with proper extension!','Error','error');
return;
end
RGB = imread(handles.fullPath);
handles.RGB = RGB;
handles.fileLoaded = 1;
set(handles.axes1,'Visible','on');
axes(handles.axes1); cla; imshow(RGB);
guidata(hObject,handles)
% --- Executes on button press in CROP1.
function CROP1_Callback(hObject, eventdata, handles)
% hObject handle to CROP1 (see GCBO)
% eventdata reserved - to be defined in a future version of MATLAB
% handles structure with handles and user data (see GUIDATA)
crop=imcrop(handles.RGB);
crop=rgb2gray(crop);
handles.crop=crop;
axes(handles.axes1);imshow(handles.crop);
guidata(hObject,handles)
% --- Executes on button press in LOAD2.
function LOAD2_Callback(hObject, eventdata, handles)
% hObject handle to LOAD2 (see GCBO)
% eventdata reserved - to be defined in a future version of MATLAB
% handles structure with handles and user data (see GUIDATA)
[FileName,PathName] = uigetfile({'*.*'},'Load Image File');
if (FileName==0) % cancel pressed
return;
end
handles.fullPath = [PathName FileName];
[a, b, Ext] = fileparts(FileName);
availableExt = {'.bmp','.jpg','.jpeg','.tiff','.png','.gif'};
FOUND = 0;
for (i=1:length(availableExt))
if (strcmpi(Ext, availableExt{i}))
FOUND=1;
break;
end
end
if (FOUND==0)
msgbox('File type not supported Load file with proper extension!','Error','error');
return;
end
RGB1= imread(handles.fullPath);
handles.RGB1 = RGB1;
handles.fileLoaded = 1;
set(handles.axes2,'Visible','on');
axes(handles.axes2); cla; imshow(RGB1);
guidata(hObject,handles)
% --- Executes on button press in CROP2.
function CROP2_Callback(hObject, eventdata, handles)
% hObject handle to CROP2 (see GCBO)
% eventdata reserved - to be defined in a future version of MATLAB
% handles structure with handles and user data (see GUIDATA)
crop1=imcrop(handles.RGB1);
crop1=rgb2gray(crop1);
handles.crop1=crop1;
axes(handles.axes2);imshow(handles.crop1);
guidata(hObject,handles)
% --- Executes on button press in TAMURA1.
function TAMURA1_Callback(hObject, eventdata, handles)
% hObject handle to TAMURA1 (see GCBO)
% eventdata reserved - to be defined in a future version of MATLAB
% handles structure with handles and user data (see GUIDATA)
%trauma features
I=im2double(handles.crop);
[Nx,Ny] = size(I);
Ng=256;
G=double(I);
abhi=zeros(Nx,Ny);
E0h=zeros(Nx,Ny);
E0v=zeros(Nx,Ny);
E1h=zeros(Nx,Ny);
E1v=zeros(Nx,Ny);
E2h=zeros(Nx,Ny);
E2v=zeros(Nx,Ny);
E3h=zeros(Nx,Ny);
E3v=zeros(Nx,Ny);
E4h=zeros(Nx,Ny);
E4v=zeros(Nx,Ny);
E5h=zeros(Nx,Ny);
E5v=zeros(Nx,Ny);
flag=0;
for i=1:Nx
for j=2:Ny
E0h(i,j)=G(i,j)-G(i,j-1);
end
end
E0h=E0h/2;
for i=1:Nx-1
for j=1:Ny
E0v(i,j)=G(i,j)-G(i+1,j);
end
end
E0v=E0v/2;
if (Nx<4||Ny<4)
flag=1;
end
if(flag==0)
for i=1:Nx-1
for j=3:Ny-1
E1h(i,j)=sum(sum(G(i:i+1,j:j+1)))-sum(sum(G(i:i+1,j-2:j-1)));
end
end
for i=2:Nx-2
for j=2:Ny
E1v(i,j)=sum(sum(G(i-1:i,j-1:j)))-sum(sum(G(i+1:i+2,j-1:j)));
end
end
E1h=E1h/4;
E1v=E1v/4;
end
if (Nx<8||Ny<8)
flag=1;
end
if(flag==0)
for i=2:Nx-2
for j=5:Ny-3
E2h(i,j)=sum(sum(G(i-1:i+2,j:j+3)))-sum(sum(G(i-1:i+2,j-4:j-1)));
end
end
for i=4:Nx-4
for j=3:Ny-1
E2v(i,j)=sum(sum(G(i-3:i,j-2:j+1)))-sum(sum(G(i+1:i+4,j-2:j+1)));
end
end
E2h=E2h/16;
E2v=E2v/16;
end
if (Nx<16||Ny<16)
flag=1;
end
if(flag==0)
for i=4:Nx-4
for j=9:Ny-7
E3h(i,j)=sum(sum(G(i-3:i+4,j:j+7)))-sum(sum(G(i-3:i+4,j-8:j-1)));
end
end
for i=8:Nx-8
for j=5:Ny-3
E3v(i,j)=sum(sum(G(i-7:i,j-4:j+3)))-sum(sum(G(i+1:i+8,j-4:j+3)));
end
end
E3h=E3h/64;
E3v=E3v/64;
end
if (Nx<32||Ny<32)
flag=1;
end
if(flag==0)
for i=8:Nx-8
for j=17:Ny-15
E4h(i,j)=sum(sum(G(i-7:i+8,j:j+15)))-sum(sum(G(i-7:i+8,j-16:j-1)));
end
end
for i=16:Nx-16
for j=9:Ny-7
E4v(i,j)=sum(sum(G(i-15:i,j-8:j+7)))-sum(sum(G(i+1:i+16,j-8:j+7)));
end
end
E4h=E4h/256;
E4v=E4v/256;
end
if (Nx<64||Ny<64)
flag=1;
end
if(flag==0)
for i=16:Nx-16
for j=33:Ny-31
E5h(i,j)=sum(sum(G(i-15:i+16,j:j+31)))-sum(sum(G(i-15:i+16,j-32:j-31)));
end
end
for i=32:Nx-32
for j=17:Ny-15
E5v(i,j)=sum(sum(G(i-31:i,j-16:j+15)))-sum(sum(G(i+1:i+32,j-16:j+15)));
end
end
E5h=E5h/1024;
E5v=E5v/1024;
end
for i=1:Nx
for j=1:Ny
[maxv,index]=max([E0h(i,j),E0v(i,j),E1h(i,j),E1v(i,j),E2h(i,j),E2v(i,j),E3h(i,j),E3v(i,j),E4h(i,j),E4v(i,j),E5h(i,j),E5v(i,j)]);
k=floor((index+1)/2);
abhi(i,j)=2.^k;
end
end
coarseness=sum(sum(abhi))/(Nx*Ny);
[counts,graylevels]=imhist(I);
PI=counts/(Nx*Ny);
averagevalue=sum(graylevels.*PI);
u4=sum((graylevels-repmat(averagevalue,[256,1])).^4.*PI);
standarddeviation=sum((graylevels-repmat(averagevalue,[256,1])).^2.*PI);
alpha4=u4/standarddeviation^2;
contrast=sqrt(standarddeviation)/alpha4.^(1/4);
PrewittH=[-1 0 1;-1 0 1;-1 0 1];
PrewittV=[1 1 1;0 0 0;-1 -1 -1];
deltaH=zeros(Nx,Ny);
for i=2:Nx-1
for j=2:Ny-1
deltaH(i,j)=sum(sum(G(i-1:i+1,j-1:j+1).*PrewittH));
end
end
for j=2:Ny-1
deltaH(1,j)=G(1,j+1)-G(1,j);
deltaH(Nx,j)=G(Nx,j+1)-G(Nx,j);
end
for i=1:Nx
deltaH(i,1)=G(i,2)-G(i,1);
deltaH(i,Ny)=G(i,Ny)-G(i,Ny-1);
end
deltaV=zeros(Nx,Ny);
for i=2:Nx-1
for j=2:Ny-1
deltaV(i,j)=sum(sum(G(i-1:i+1,j-1:j+1).*PrewittV));
end
end
for j=1:Ny
deltaV(1,j)=G(2,j)-G(1,j);
deltaV(Nx,j)=G(Nx,j)-G(Nx-1,j);
end
for i=2:Nx-1
deltaV(i,1)=G(i+1,1)-G(i,1);
deltaV(i,Ny)=G(i+1,Ny)-G(i,Ny);
end
deltaG=(abs(deltaH)+abs(deltaV))/2;
theta=zeros(Nx,Ny);
for i=1:Nx
for j=1:Ny
if (deltaH(i,j)==0)&&(deltaV(i,j)==0)
elseif deltaH(i,j)==0
theta(i,j)=pi;
else
theta(i,j)=atan(deltaV(i,j)/deltaH(i,j))+pi/2;
end
end
end
theta1=reshape(theta,1,[]);
phai=0:0.0001:pi;
HD1=hist(theta1,phai);
HD1=HD1/(Nx*Ny);
HD2=zeros(size(HD1));
THRESHOLD=0;
for m=1:length(HD2)
if HD1(m)>=THRESHOLD
HD2(m)=HD1(m);
end
end
[c,index]=max(HD2);
phaiP=index*0.0001;
direction=0;
for m=1:length(HD2)
if HD2(m)~=0
direction=direction+(phai(m)-phaiP)^2*HD2(m);
end
end
disp('Trauma features _Coarseness');display(coarseness)
disp('Trauma features _Contrast');display(contrast)
disp('Trauma features _Direction');display(direction)
set(handles.edit1,'String',coarseness);
set(handles.edit2,'String',contrast);
set(handles.edit5,'String',direction);
guidata(hObject,handles)
% --- Executes on button press in TAMURA2.
function TAMURA2_Callback(hObject, eventdata, handles)
% hObject handle to TAMURA2 (see GCBO)
% eventdata reserved - to be defined in a future version of MATLAB
% handles structure with handles and user data (see GUIDATA)
I=im2double(handles.crop1);
[Nx,Ny] = size(I);
Ng=256;
G=double(I);
abhi=zeros(Nx,Ny);
E0h=zeros(Nx,Ny);
E0v=zeros(Nx,Ny);
E1h=zeros(Nx,Ny);
E1v=zeros(Nx,Ny);
E2h=zeros(Nx,Ny);
E2v=zeros(Nx,Ny);
E3h=zeros(Nx,Ny);
E3v=zeros(Nx,Ny);
E4h=zeros(Nx,Ny);
E4v=zeros(Nx,Ny);
E5h=zeros(Nx,Ny);
E5v=zeros(Nx,Ny);
flag=0;
for i=1:Nx
for j=2:Ny
E0h(i,j)=G(i,j)-G(i,j-1);
end
end
E0h=E0h/2;
for i=1:Nx-1
for j=1:Ny
E0v(i,j)=G(i,j)-G(i+1,j);
end
end
E0v=E0v/2;
if (Nx<4||Ny<4)
flag=1;
end
if(flag==0)
for i=1:Nx-1
for j=3:Ny-1
E1h(i,j)=sum(sum(G(i:i+1,j:j+1)))-sum(sum(G(i:i+1,j-2:j-1)));
end
end
for i=2:Nx-2
for j=2:Ny
E1v(i,j)=sum(sum(G(i-1:i,j-1:j)))-sum(sum(G(i+1:i+2,j-1:j)));
end
end
E1h=E1h/4;
E1v=E1v/4;
end
if (Nx<8||Ny<8)
flag=1;
end
if(flag==0)
for i=2:Nx-2
for j=5:Ny-3
E2h(i,j)=sum(sum(G(i-1:i+2,j:j+3)))-sum(sum(G(i-1:i+2,j-4:j-1)));
end
end
for i=4:Nx-4
for j=3:Ny-1
E2v(i,j)=sum(sum(G(i-3:i,j-2:j+1)))-sum(sum(G(i+1:i+4,j-2:j+1)));
end
end
E2h=E2h/16;
E2v=E2v/16;
end
if (Nx<16||Ny<16)
flag=1;
end
if(flag==0)
for i=4:Nx-4
for j=9:Ny-7
E3h(i,j)=sum(sum(G(i-3:i+4,j:j+7)))-sum(sum(G(i-3:i+4,j-8:j-1)));
end
end
for i=8:Nx-8
for j=5:Ny-3
E3v(i,j)=sum(sum(G(i-7:i,j-4:j+3)))-sum(sum(G(i+1:i+8,j-4:j+3)));
end
end
E3h=E3h/64;
E3v=E3v/64;
end
if (Nx<32||Ny<32)
flag=1;
end
if(flag==0)
for i=8:Nx-8
for j=17:Ny-15
E4h(i,j)=sum(sum(G(i-7:i+8,j:j+15)))-sum(sum(G(i-7:i+8,j-16:j-1)));
end
end
for i=16:Nx-16
for j=9:Ny-7
E4v(i,j)=sum(sum(G(i-15:i,j-8:j+7)))-sum(sum(G(i+1:i+16,j-8:j+7)));
end
end
E4h=E4h/256;
E4v=E4v/256;
end
if (Nx<64||Ny<64)
flag=1;
end
if(flag==0)
for i=16:Nx-16
for j=33:Ny-31
E5h(i,j)=sum(sum(G(i-15:i+16,j:j+31)))-sum(sum(G(i-15:i+16,j-32:j-31)));
end
end
for i=32:Nx-32
for j=17:Ny-15
E5v(i,j)=sum(sum(G(i-31:i,j-16:j+15)))-sum(sum(G(i+1:i+32,j-16:j+15)));
end
end
E5h=E5h/1024;
E5v=E5v/1024;
end
for i=1:Nx
for j=1:Ny
[maxv,index]=max([E0h(i,j),E0v(i,j),E1h(i,j),E1v(i,j),E2h(i,j),E2v(i,j),E3h(i,j),E3v(i,j),E4h(i,j),E4v(i,j),E5h(i,j),E5v(i,j)]);
k=floor((index+1)/2);
abhi(i,j)=2.^k;
end
end
coarseness=sum(sum(abhi))/(Nx*Ny);
[counts,graylevels]=imhist(I);
PI=counts/(Nx*Ny);
averagevalue=sum(graylevels.*PI);
u4=sum((graylevels-repmat(averagevalue,[256,1])).^4.*PI);
standarddeviation=sum((graylevels-repmat(averagevalue,[256,1])).^2.*PI);
alpha4=u4/standarddeviation^2;
contrast=sqrt(standarddeviation)/alpha4.^(1/4);
PrewittH=[-1 0 1;-1 0 1;-1 0 1];
PrewittV=[1 1 1;0 0 0;-1 -1 -1];
deltaH=zeros(Nx,Ny);
for i=2:Nx-1
for j=2:Ny-1
deltaH(i,j)=sum(sum(G(i-1:i+1,j-1:j+1).*PrewittH));
end
end
for j=2:Ny-1
deltaH(1,j)=G(1,j+1)-G(1,j);
deltaH(Nx,j)=G(Nx,j+1)-G(Nx,j);
end
for i=1:Nx
deltaH(i,1)=G(i,2)-G(i,1);
deltaH(i,Ny)=G(i,Ny)-G(i,Ny-1);
end
deltaV=zeros(Nx,Ny);
for i=2:Nx-1
for j=2:Ny-1
deltaV(i,j)=sum(sum(G(i-1:i+1,j-1:j+1).*PrewittV));
end
end
for j=1:Ny
deltaV(1,j)=G(2,j)-G(1,j);
deltaV(Nx,j)=G(Nx,j)-G(Nx-1,j);
end
for i=2:Nx-1
deltaV(i,1)=G(i+1,1)-G(i,1);
deltaV(i,Ny)=G(i+1,Ny)-G(i,Ny);
end
deltaG=(abs(deltaH)+abs(deltaV))/2;
theta=zeros(Nx,Ny);
for i=1:Nx
for j=1:Ny
if (deltaH(i,j)==0)&&(deltaV(i,j)==0)
elseif deltaH(i,j)==0
theta(i,j)=pi;
else
theta(i,j)=atan(deltaV(i,j)/deltaH(i,j))+pi/2;
end
end
end
theta1=reshape(theta,1,[]);
phai=0:0.0001:pi;
HD1=hist(theta1,phai);
HD1=HD1/(Nx*Ny);
HD2=zeros(size(HD1));
THRESHOLD=0;
for m=1:length(HD2)
if HD1(m)>=THRESHOLD
HD2(m)=HD1(m);
end
end
[c,index]=max(HD2);
phaiP=index*0.0001;
direction=0;
for m=1:length(HD2)
if HD2(m)~=0
direction=direction+(phai(m)-phaiP)^2*HD2(m);
end
end
disp('Trauma features _Coarseness');display(coarseness)
disp('Trauma features _Contrast');display(contrast)
disp('Trauma features _Direction');display(direction)
set(handles.edit9,'String',coarseness);
set(handles.edit10,'String',contrast);
set(handles.edit11,'String',direction);
guidata(hObject,handles)
function edit1_Callback(hObject, eventdata, handles)
% hObject handle to edit1 (see GCBO)
% eventdata reserved - to be defined in a future version of MATLAB
% handles structure with handles and user data (see GUIDATA)
% Hints: get(hObject,'String') returns contents of edit1 as text
% str2double(get(hObject,'String')) returns contents of edit1 as a double
% --- Executes during object creation, after setting all properties.
function edit1_CreateFcn(hObject, eventdata, handles)
% hObject handle to edit1 (see GCBO)
% eventdata reserved - to be defined in a future version of MATLAB
% handles empty - handles not created until after all CreateFcns called
% Hint: edit controls usually have a white background on Windows.
% See ISPC and COMPUTER.
if ispc && isequal(get(hObject,'BackgroundColor'), get(0,'defaultUicontrolBackgroundColor'))
set(hObject,'BackgroundColor','white');
end
function edit2_Callback(hObject, eventdata, handles)
% hObject handle to edit2 (see GCBO)
% eventdata reserved - to be defined in a future version of MATLAB
% handles structure with handles and user data (see GUIDATA)
% Hints: get(hObject,'String') returns contents of edit2 as text
% str2double(get(hObject,'String')) returns contents of edit2 as a double
% --- Executes during object creation, after setting all properties.
function edit2_CreateFcn(hObject, eventdata, handles)
% hObject handle to edit2 (see GCBO)
% eventdata reserved - to be defined in a future version of MATLAB
% handles empty - handles not created until after all CreateFcns called
% Hint: edit controls usually have a white background on Windows.
% See ISPC and COMPUTER.
if ispc && isequal(get(hObject,'BackgroundColor'), get(0,'defaultUicontrolBackgroundColor'))
set(hObject,'BackgroundColor','white');
end
function edit5_Callback(hObject, eventdata, handles)
% hObject handle to edit5 (see GCBO)
% eventdata reserved - to be defined in a future version of MATLAB
% handles structure with handles and user data (see GUIDATA)
% Hints: get(hObject,'String') returns contents of edit5 as text
% str2double(get(hObject,'String')) returns contents of edit5 as a double
% --- Executes during object creation, after setting all properties.
function edit5_CreateFcn(hObject, eventdata, handles)
% hObject handle to edit5 (see GCBO)
% eventdata reserved - to be defined in a future version of MATLAB
% handles empty - handles not created until after all CreateFcns called
% Hint: edit controls usually have a white background on Windows.
% See ISPC and COMPUTER.
if ispc && isequal(get(hObject,'BackgroundColor'), get(0,'defaultUicontrolBackgroundColor'))
set(hObject,'BackgroundColor','white');
end
function edit6_Callback(hObject, eventdata, handles)
% hObject handle to edit6 (see GCBO)
% eventdata reserved - to be defined in a future version of MATLAB
% handles structure with handles and user data (see GUIDATA)
% Hints: get(hObject,'String') returns contents of edit6 as text
% str2double(get(hObject,'String')) returns contents of edit6 as a double
% --- Executes during object creation, after setting all properties.
function edit6_CreateFcn(hObject, eventdata, handles)
% hObject handle to edit6 (see GCBO)
% eventdata reserved - to be defined in a future version of MATLAB
% handles empty - handles not created until after all CreateFcns called
% Hint: edit controls usually have a white background on Windows.
% See ISPC and COMPUTER.
if ispc && isequal(get(hObject,'BackgroundColor'), get(0,'defaultUicontrolBackgroundColor'))
set(hObject,'BackgroundColor','white');
end
function edit7_Callback(hObject, eventdata, handles)
% hObject handle to edit7 (see GCBO)
% eventdata reserved - to be defined in a future version of MATLAB
% handles structure with handles and user data (see GUIDATA)
% Hints: get(hObject,'String') returns contents of edit7 as text
% str2double(get(hObject,'String')) returns contents of edit7 as a double
% --- Executes during object creation, after setting all properties.
function edit7_CreateFcn(hObject, eventdata, handles)
% hObject handle to edit7 (see GCBO)
% eventdata reserved - to be defined in a future version of MATLAB
% handles empty - handles not created until after all CreateFcns called
% Hint: edit controls usually have a white background on Windows.
% See ISPC and COMPUTER.
if ispc && isequal(get(hObject,'BackgroundColor'), get(0,'defaultUicontrolBackgroundColor'))
set(hObject,'BackgroundColor','white');
end
function edit8_Callback(hObject, eventdata, handles)
% hObject handle to edit8 (see GCBO)
% eventdata reserved - to be defined in a future version of MATLAB
% handles structure with handles and user data (see GUIDATA)
% Hints: get(hObject,'String') returns contents of edit8 as text
% str2double(get(hObject,'String')) returns contents of edit8 as a double
% --- Executes during object creation, after setting all properties.
function edit8_CreateFcn(hObject, eventdata, handles)
% hObject handle to edit8 (see GCBO)
% eventdata reserved - to be defined in a future version of MATLAB
% handles empty - handles not created until after all CreateFcns called
% Hint: edit controls usually have a white background on Windows.
% See ISPC and COMPUTER.
if ispc && isequal(get(hObject,'BackgroundColor'), get(0,'defaultUicontrolBackgroundColor'))
set(hObject,'BackgroundColor','white');
end
function edit9_Callback(hObject, eventdata, handles)
% hObject handle to edit9 (see GCBO)
% eventdata reserved - to be defined in a future version of MATLAB
% handles structure with handles and user data (see GUIDATA)
% Hints: get(hObject,'String') returns contents of edit9 as text
% str2double(get(hObject,'String')) returns contents of edit9 as a double
% --- Executes during object creation, after setting all properties.
function edit9_CreateFcn(hObject, eventdata, handles)
% hObject handle to edit9 (see GCBO)
% eventdata reserved - to be defined in a future version of MATLAB
% handles empty - handles not created until after all CreateFcns called
% Hint: edit controls usually have a white background on Windows.
% See ISPC and COMPUTER.
if ispc && isequal(get(hObject,'BackgroundColor'), get(0,'defaultUicontrolBackgroundColor'))
set(hObject,'BackgroundColor','white');
end
function edit10_Callback(hObject, eventdata, handles)
% hObject handle to edit10 (see GCBO)
% eventdata reserved - to be defined in a future version of MATLAB
% handles structure with handles and user data (see GUIDATA)
% Hints: get(hObject,'String') returns contents of edit10 as text
% str2double(get(hObject,'String')) returns contents of edit10 as a double
% --- Executes during object creation, after setting all properties.
function edit10_CreateFcn(hObject, eventdata, handles)
% hObject handle to edit10 (see GCBO)
% eventdata reserved - to be defined in a future version of MATLAB
% handles empty - handles not created until after all CreateFcns called
% Hint: edit controls usually have a white background on Windows.
% See ISPC and COMPUTER.
if ispc && isequal(get(hObject,'BackgroundColor'), get(0,'defaultUicontrolBackgroundColor'))
set(hObject,'BackgroundColor','white');
end
function edit11_Callback(hObject, eventdata, handles)
% hObject handle to edit11 (see GCBO)
% eventdata reserved - to be defined in a future version of MATLAB
% handles structure with handles and user data (see GUIDATA)
% Hints: get(hObject,'String') returns contents of edit11 as text
% str2double(get(hObject,'String')) returns contents of edit11 as a double
% --- Executes during object creation, after setting all properties.
function edit11_CreateFcn(hObject, eventdata, handles)
% hObject handle to edit11 (see GCBO)
% eventdata reserved - to be defined in a future version of MATLAB
% handles empty - handles not created until after all CreateFcns called
% Hint: edit controls usually have a white background on Windows.
% See ISPC and COMPUTER.
if ispc && isequal(get(hObject,'BackgroundColor'), get(0,'defaultUicontrolBackgroundColor'))
set(hObject,'BackgroundColor','white');
end